Prof. Upinder S. Bhalla - Protein Kinase C
Protein Kinase C modeling example
PKC activation
PKC is activated by Ca, AA, and DAG synergistically. We matched experimental concentration-effect curves from two main sources:
- J. D. Schaechter and L. I. Benowitz, J. Neurosci. 13, 4361 (1993);
- T. Shinomura, Y. Asaoka, M. Oka, K. Yoshida, Y. Nishizuka, Proc. Natl. Acad. Sci. U.S.A. 88, 5149 (1991)
Our simulations were built up iteratively: First we matched AA activation of PKC at zero Ca. Then we matched activation of PKC with Ca at zero AA. We then matched the curves in B with 1 uM Ca and varying AA. At this stage we were able to test the match for C, with varying Ca and 50 uM AA. The match was excellent. Finally, we incorporated DAG interactions into the model.
Each time we included additional interactions the parameters for the previously matched values had to be revised to ensure that the original curve-fits remained valid.
The curve matches are well within the range of experimental error, especially when the variations reported between different groups are considered.
A. Block diagram of activation for PKC pathway by Ca2+, AA and DAG.
B: Activation of PKC by AA, with (triangles) or without (squares) 1 mM Ca2+.
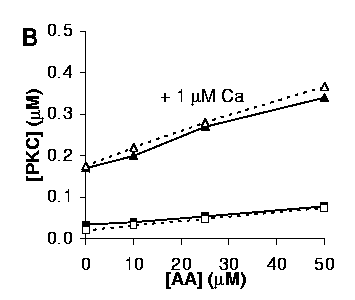
Open symbols and dashed lines represent simulations, solid symbols and solid lines are experimental data.
C: Activation of PKC by Ca2+, with (triangles) or without (squares) 50 mM AA.
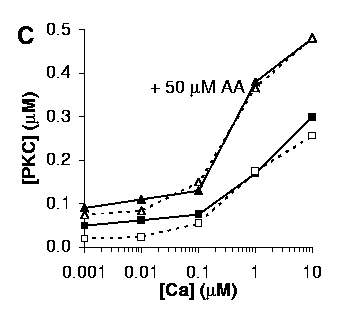
The curve in the presence of 50 mM AA (triangles) was predicted from the parameters obtained by matching the curves in B and the curve without AA (squares) in C, without further adjustment.
D: Activation of PKC by DAG, with (triangles) or without (squares) 50 mM AA.
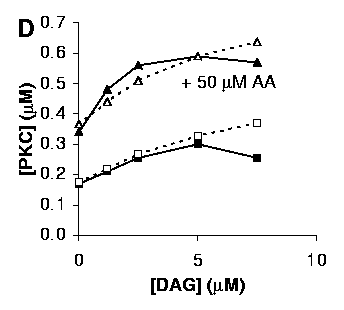
Both curves in D were obtained in the presence of 1 mM Ca2+. Due to different methods for estimating DAG concentrations the levels of DAG used in the model are scaled 15-fold up with respect to the experimental conditions from Shinomura et al.



