Dr Yamuna Krishnan - Research
Structure and Dynamics of Nucleic Acids
What drives tetramerization in the I-motif?
Souvik Modi and Saikat Chakraborty
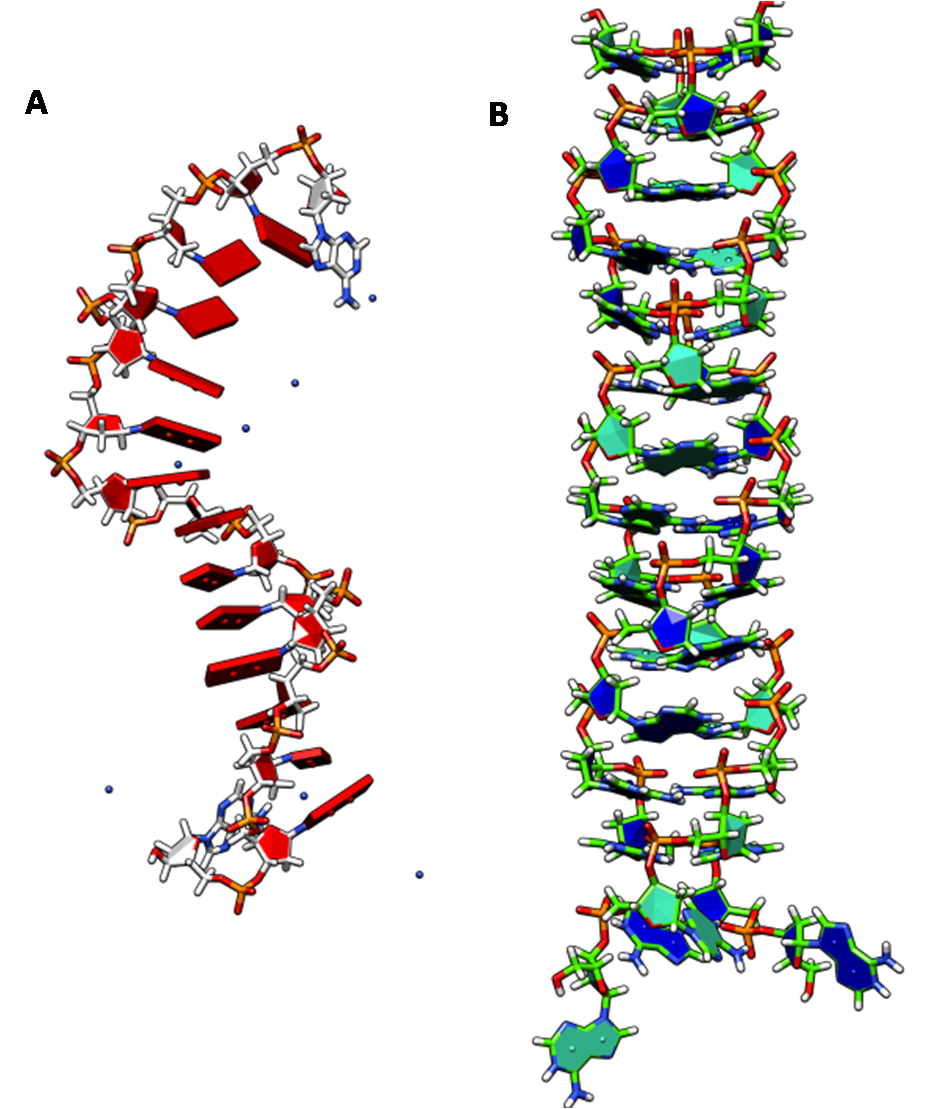
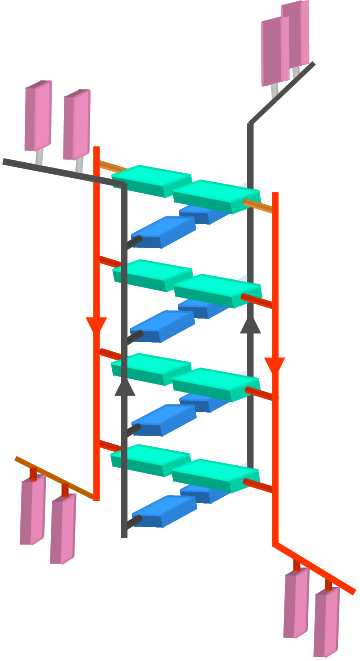
The I-motif is a four-stranded nucleic acid structure formed from C-rich sequences and consists of two parallel stranded duplexes formed from hemi-protonated cytosines that intercalate (Figure 2). One of the prevailing issues regarding DNA i-motifs has been in understanding why intercalation should occur at all. In a natural DNA i-motif, duplex intercalation results in a remarkably narrow groove that positions two negatively charged DNA backbones extremely close resulting in greater electrostatic repulsion. Sugar-sugar contacts along the backbones of both strands that flank the narrow grooves of the i-motif have been implicated in tetramer stabilization although evidence for the contrary also exists. We have shown that hybrid i-motifs are an excellent platform to probe interactions in the narrow groove. It is the only method that can identify molecular interactions that promote or ‘positive regulate’ i-motif formation. All other methods have identified only destabilisers of i-motifs. Our work has shown that (i) sugar-sugar contacts in i-motifs are not a consequence of, but possibly drive tetramerization (ii) 2’OH steric clash in the narrow groove is not as destabilizing as perceived (iii) i-motif intercalation topology is highly sensitive to narrow groove interactions and (iv) there is a subtle specificity encoded even in the sugar-sugar contacts.
DNA nanomachine that maps spatial and temporal pH inside living cells.
Souvik Modi, Swetha MG
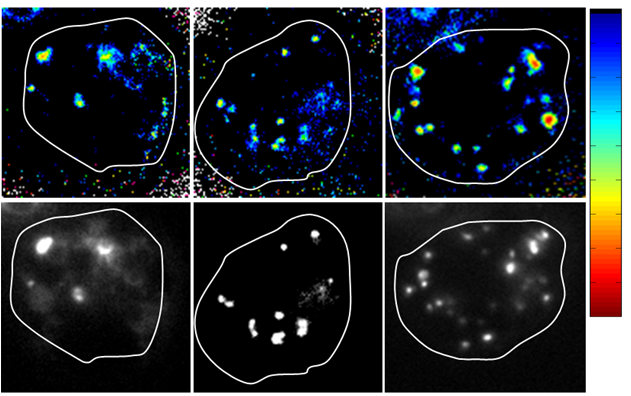
DNA nanomachines are synthetic DNA assemblies that switch between defined molecular conformations upon stimulation by external triggers. Hitherto, the performance of DNA devices has been limited to in vitro applications. Here we report the construction of a DNA nanomachine triggered by protons, called the I-switch that functions as a FRET-based pH sensor inside living cells. It is an efficient reporter of pH from 5.5 to 7, with high dynamic range between pH 5.8 -7. To demonstrate its ability to function inside living cells we use the I-switch to map spatial and temporal pH changes associated with endosome maturation. Thus the performance of our DNA nanodevices inside living systems illustrates the potential of DNA scaffolds responsive to more complex triggers in sensing, diagnostics and targeted therapies in living systems
Collaborators: Dr. Satyajit Mayor (NCBS)
The Poly dA helix: a new structural motif for high performance DNA-based molecular switches
Saikat Chakraborty, Suruchi Sharma
We report a pH-dependent conformational transition in short,defined homopolymeric deoxyadenosines (dA15) from a single helicalstructure with stacked nucleobases at neutral pH to a double-helical,parallel-stranded duplex held together by AH+-H+A base pairsat acidic pH. Using native PAGE, 2D NMR, circular dichroism(CD) and fluorescence spectroscopy, we have characterized thetwo different pH dependent forms of dA15. The pH-triggered transitionbetween the two defined helical forms of dA15 is characterizedby CD and fluorescence. The kinetics of this conformationalswitch is found to occur on a millisecond time scale. This robust,highly reversible, pH-induced transition between the two well-definedstructured states of dA15 represents a new molecular buildingblock for the construction of quick-response, pH-switchablearchitectures in structural DNA nanotechnology.
Collaborator: Dr. Prabal Maiti, Physic Dept. IISc