Dr. Ranabir Das
Laboratory of Biomolecular Structure, Dynamics and Function
Research Interests
Host-Microbe Interactions
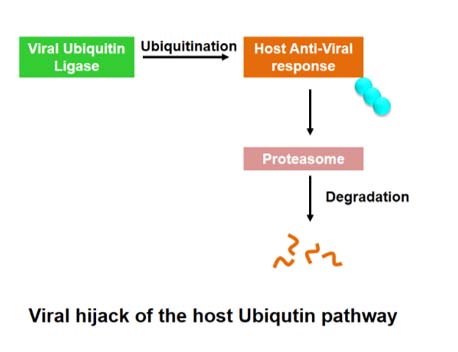
Microbes hijack the host machinery to their own advantage. During infection, several host defense mechanisms are destabilized by the microbes using the ubiquitin-proteasome pathway. We study the interactions between microbial factors and host proteins to understand this process.
Protein Quality Control
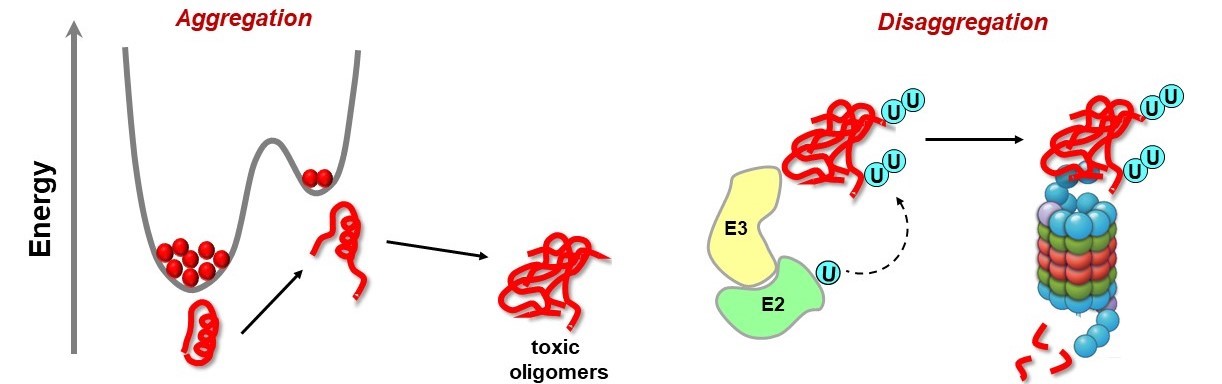
Misfolding of proteins can lead to accimulation of toxic aggregates. It is assumed that higher energy intermediates along the protein folding landscape seed the formation of toxic aggregates. Protein quality control systems like the Ubiquitin-proteasome pathway ensures that these potentially toxic species are degraded. We are interested to get insights into the structure of higher energy intermediates. We also investigate how misfolded proteins targeted by the ubiquitin-proteasome pathway.
Positions: We are actively looking for postdocs and CSIR PhD fellows interested in studying Biophysics and Biochemistry of host-pathogen interactions and proteinopathies. Please apply with a detailed CV to rana@ncbs.res.in. Your CV should reflect your expertise and research interest.
Students can also to apply through the NCBS-PhD program. There are no summer/winter positions available.



