What makes plants grow? One might focus on the soil quality or other environmental conditions, but there is a vital in-built aspect that interacts with these for plant growth: the genome, consisting of all the genetic information the organism carries. People and plants, like many living organisms, have genes. But there are differences in the genetic material we carry, as well as the way these get expressed from person to person or plant to plant. Gene expression is the manner in which proteins are produced, based on the instructions encoded in an organism’s genes. We know that genes get passed on from parents to offspring, this applies to plants and humans.
In the Epigenetics Lab at NCBS, led by Dr. P. V. Shivaprasad, the modulation or regulation of plant genomes is at the heart of the research. They study how initiating and maintaining heritable changes in gene expression can happen, without any changes in the DNA sequence of genes. These epigenetic changes are influenced by the environmental factors around the organism, and there are many layers of regulation.
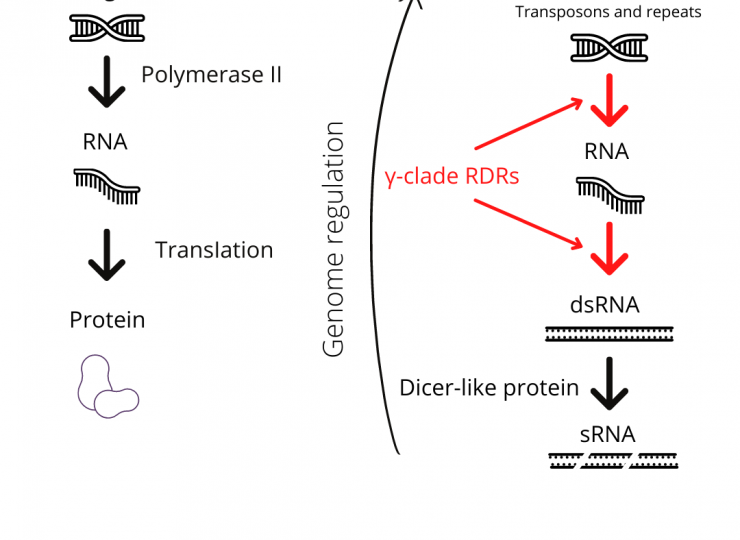
One of the levels of regulation is from small (s)RNAs. These regulators play a major role in the establishment and maintenance of epigenetic marks. sRNAs are master regulators of gene expression, involved in a multitude of functions in development, resistance, and genome integrity in plants and most organisms. But the production of sRNAs is controlled by several proteins in the plant genome. These typically include polymerases and Dicers, along with their partner proteins.
In a recent research article published in New Phytologist, researchers from the lab have shown the role of a class of enzymes—γ-clade RNA-dependent RNA polymerases (RDRs)—in the development and yield-related traits of rice, through the regulation of sRNAs and other RNAs.
RNA silencing is a regulatory mechanism that employs the sRNAs and associated effector proteins to induce silencing at transcriptional or post-transcriptional levels of the gene. Double-stranded (ds)RNAs are the trigger for gene silencing in plants and a precursor to sRNAs, and in this study, the researchers examine how dsRNA generation is dependent on the γ-clade RDRs.
This is the first evidence of the importance of γ-clade RNA-dependent RNA polymerases in plant development, their atypical biochemical activities, and their contribution to the regulation of gene expression. These enzymes are unusual in that they are found only in plants and fungi which are considered genetically distant from each other. These new players in genome regulation are the ones that keep some unruly elements in the genome under check, so that when they work well, plants develop faster and produce more seeds. If they are absent, the genome cannot decide what region is important and the plants are unable to grow properly.
“Our results highlight a previously unknown mechanism of genome regulation involving a new set of regulators named γ-clade RDRs. It is surprising that these enzymes can transcribe nascent RNAs and also convert them to dsRNAs. As seen from our genetic studies, these proteins are very important for the growth and development of rice and they can be important candidates for further enhancing the yield-related traits in the crop plants,” says Vikram Jha, lead author of the study.